1D Data - 1D Inversion
[1]:
# This file is part of ILTpy examples.
# Author : Dr. Davis Thomas Daniel
# Last updated : 16.02.2026
This example shows how to do a 1D inversion using ILTPy.
For this example, we will first simulate some synthetic data using Kohlrausch–Williams–Watts function with some postive and negative peaks.
This example was run on a machine with Apple M1 processor (8 core CPU) and 16 GB of memory.
Imports
[2]:
import iltpy as ilt
import numpy as np
import matplotlib.pyplot as plt
print(f"ILTpy version: {ilt.__version__}")
ILTpy version: 1.1.0
Generating synthetic data
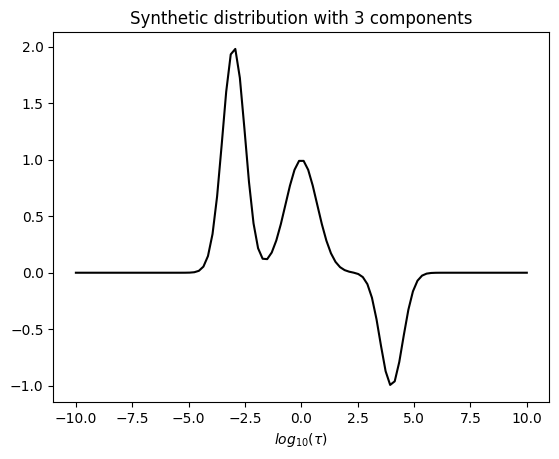
The true distribution
[3]:
def gaussian(x, mu, sigma, amplitude):
return amplitude * np.exp(-0.5 * ((x - mu) / sigma) ** 2)
# Generate synthetic spectrum
tau = np.linspace(-10, 10, 100)
components = [-3, 0, 4]
sigmas = [0.5, 0.7, 0.5]
amplitudes = [2,1,-1]
dist1d = np.zeros_like(tau)
# Add multiple Gaussian components to the spectrum
for mu, sigma, amplitude in zip(components, sigmas, amplitudes):
dist1d += gaussian(tau, mu, sigma, amplitude)
Visualize the true distribution
[4]:
# Plot the result
plt.plot(tau, dist1d,'k')
plt.title('Synthetic distribution with 3 components')
plt.xlabel(r'$log_{10}(\tau)$')
plt.show()
Generate time-domain data
[5]:
## Time domain data :
#timing
t_syn = np.logspace(-10,10,1024).reshape((-1,1))
tau = np.linspace(-10, 10, 100).reshape((1,-1))
# kernel matrix
K_matrix = ilt.Exponential().kernel(t_syn,10**tau)
snr = 100
syn_data_1D = K_matrix @ dist1d
scaling_factor = snr/np.std(syn_data_1D)
syn_data_1D*=scaling_factor
## add noise
np.random.seed(2505)
syn_data_1D += np.random.randn(len(t_syn))
Iltpy Workflow
Load data
[6]:
# Load the data into iltpy using iltload
ilt_1d = ilt.iltload(data=syn_data_1D, t=t_syn)
Initialize
[7]:
# Specify parameters and initialize inversion
tau = np.logspace(-10, 10, 100) # output sampling points
ilt_1d.init(tau=tau, kernel=ilt.Exponential())
Invert
[9]:
ilt_1d.invert()
Starting iterations ...
100%|██████████| 100/100 [00:00<00:00, 459.40it/s, Convergence=5.18e-04]
Done.
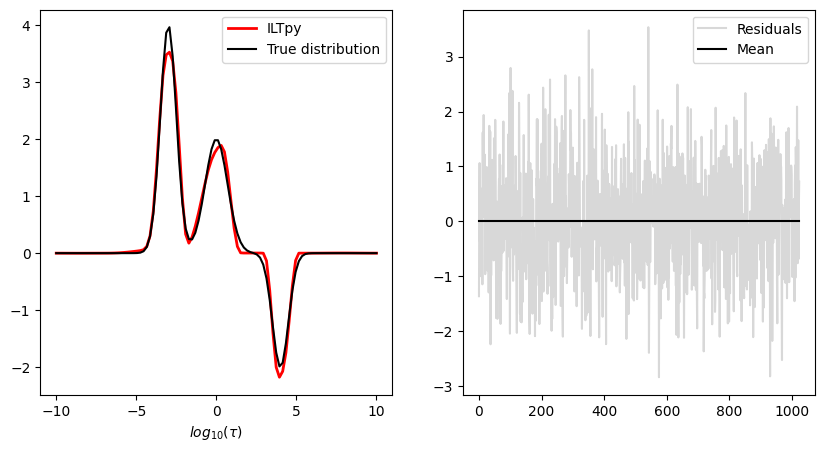
Plot results
[10]:
fig,ax = plt.subplots(ncols=2,figsize=[10,5])
ax[0].plot(np.log10(tau),dist1d*scaling_factor,label='True distribution',color='black')
ax[0].plot(np.log10(tau),ilt_1d.g,label='ILTpy',color='red',linewidth=2,linestyle="dashed")
ax[0].set_xlabel(r'$log_{10}(\tau)$')
ax[0].legend()
ax[1].plot(ilt_1d.residuals,label='Residuals',alpha=0.3,color='grey')
ax[1].plot([ilt_1d.residuals.mean()]*1024,label='Mean',color='black')
ax[1].legend()
[10]:
<matplotlib.legend.Legend at 0x10de4d7f0>
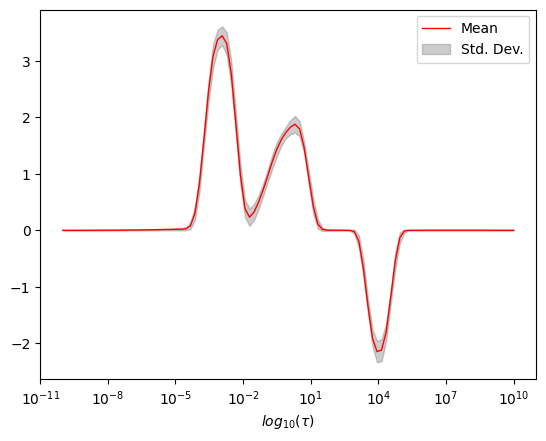
Appendix : Uncertainity Estimation
iltstatscan be used to estimate uncertainty by refitting the initial fit of the data with randomly generated noise vectors and invertingntimes.To use
iltstats, data must be inverted at least once.ncontrols the number of data samples which will be generated andn_jobscontrols the number of parallel jobs to create.
[11]:
ilt_1d.iltstats(n=200,n_jobs=-1) # n_jobs = -1 uses all available processors.
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers.
[Parallel(n_jobs=-1)]: Done 34 tasks | elapsed: 1.2s
[Parallel(n_jobs=-1)]: Done 184 tasks | elapsed: 4.4s
[Parallel(n_jobs=-1)]: Done 200 out of 200 | elapsed: 4.6s finished
Plot results
[12]:
fig,ax = plt.subplots()
ax.semilogx(ilt_1d.tau[0].flatten(),ilt_1d.statistics['g_mean'],color='r',linewidth=1,label='Mean')
ax.fill_between(ilt_1d.tau[0].flatten(),*[i for i in ilt_1d.statistics['g_conf_interval']],color='grey',alpha=0.4,label='Std. Dev.')
ax.legend()
_=ax.set_xlabel(r'$log_{10}(\tau)$')
#plt.xlim(1e4,2e6)
#_=plt.ylim(-1,22)