Bruker NMR data, nmrglue and ILTpy
[1]:
# This file is part of ILTpy examples.
# Author : Dr. Davis Thomas Daniel
# Last updated : 16.02.2026
This example shows an ILTpy workflow involving NMR data acuqired on a Bruker spectrometer, reading and processing it using nmrglue and then ILT analysis using ILTpy.
It is recommended to familiarize yourself with the first example in the NMR section before continuing.
nmrglue is a python module for reading, writing, and interacting with the spectral data stored in a number of common NMR data formats. For more information see nmrglue’s documentation.
The dataset is a 7Li inversion recovery dataset, the sample is a solid electrolyte. The measurement was done at static conditions.
Imports
[2]:
import nmrglue as ng # nmrglue
import iltpy as ilt # iltpy
import numpy as np # numpy
import matplotlib.pyplot as plt # matplotlib
print("nmrglue version: ",ng.__version__)
print("iltpy version: ",ilt.__version__)
nmrglue version: 0.11
iltpy version: 1.1.0
Read the Bruker data using nmrglue
[3]:
# read in the bruker formatted data
dic, data = ng.bruker.read('../../../examples/NMR/bruker_data_IR/14') # experiment folder 9
# remove the digital filter
data = ng.bruker.remove_digital_filter(dic, data)
# process the spectrum. This may vary for your dataset, therefore, choose appropriate values.
#data = ng.proc_base.zf_size(data, 16384) # zero fill points
data = ng.proc_base.fft(data) # Fourier transform
data = ng.proc_base.ps(data, p0=150,p1=10) # phase correction
data = ng.proc_base.di(data) # discard the imaginaries
# load the vdlist (this file is usually found in the experiment folder)
vdlist = np.loadtxt("../../../examples/NMR/bruker_data_IR/14/vdlist") # vdlist contains the recovery delays in seconds.
[4]:
print(f"Data shape : {data.shape}") # there are 11 recovery delays and each spectrum has 33202 points
Data shape : (11, 33202)
[5]:
# calculate chemical shift axis in ppm, (Important! : Reference it appropriately for your dataset)
offset = (float(dic['acqus']['SW']) / 2) - (float(dic['acqus']['O1']) / float(dic['acqus']['BF1'])) #calculate frequency offset
start = float(dic['acqus']['SW']) - offset #calculate higher limit of ppm scale
end = -offset #calculate lower limit of ppm scale
step = float(dic['acqus']['SW']) / data.shape[1] #calculate interval
chemical_shift_ppm = np.arange(start, end, -step)
print(f"Spacing of chemical shift axis : {np.mean(np.diff(chemical_shift_ppm))}")
Spacing of chemical shift axis : -0.0060528671508279785
[6]:
# The size of the first dimension (11 in this case) should match the number of delays in the vdlist
print(f"Number of delays in vdlist : {len(vdlist)}")
Number of delays in vdlist : 11
[11]:
# plot the last recovery delay
fig,ax = plt.subplots(figsize=(5,4))
ax.plot(data[-1][9000:25000]) # a region from the full spectrum
ax.set_title("7Li Spectrum (Last recovery delay)",fontsize=12)
[11]:
Text(0.5, 1.0, '7Li Spectrum (Last recovery delay)')
[12]:
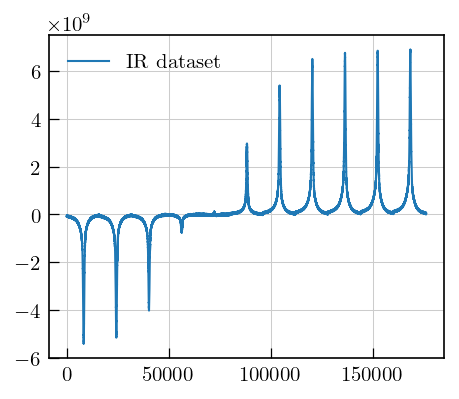
# plot the whole dataset
fig,ax = plt.subplots(figsize=(5,4))
ax.plot(data[0:11,9000:25000].flatten(),label="IR dataset") # plot all spectra as a flattened array
ax.legend()
[12]:
<matplotlib.legend.Legend at 0x125514ec0>
Data preparation
Data reduction
[13]:
# reduce the data, by reducing the sampling, this reduces the computational expense of the inversion
data_red = data.copy()
data_red = data_red[:,9000:25000:15] # we pick every 15th point from data point index 9000 to 25000
[14]:
data_red.shape
[14]:
(11, 1067)
Set the noise variance to unity
[16]:
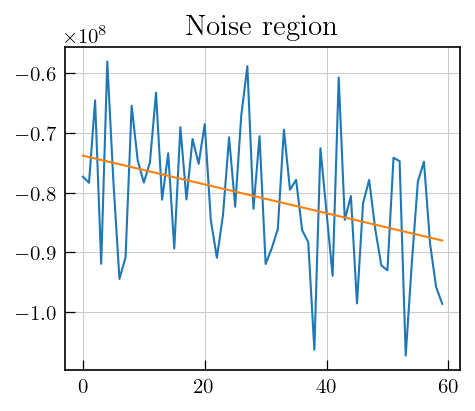
# select a region with noise, calculate standard deviation
fig,ax = plt.subplots(figsize=(5,4))
ax.plot(data_red[0][10:70]) # Here, noise region from spectrum corresponding to first recovery delay is chosen
ax.set_title("Noise region")
# fit a straight line to remove background
m,b = np.polyfit(np.arange(10,70),data_red[0][10:70],deg=1)
fit = m*np.arange(10,70)+b
plt.plot(m*np.arange(10,70)+b)
print(f"SD : {np.std(data_red[0][10:70]-fit)}") # subtract the fit and calculate standard deviation of the
SD : 10240545.292070072
[17]:
# scale the data with the obtained value from the previous step.
data_red = data_red/10240545.292070072
ILTpy workflow
Load the data
[18]:
# provide the reduced data and the vdlist to iltpy
ir_ilt = ilt.iltload(data=data_red,t=vdlist)
Initialize and invert
[19]:
# initialize by providing tau and specifying kernel
tau = np.logspace(-2.5,5,80) # output sampling vector
# specify an exponential kernel for T1, Inversion recovery
# we also specify a more closely spaced array in the non-inverted, but regularized second dimension.
ir_ilt.init(tau,kernel=ilt.Exponential(),dim_ndim=np.arange(1,1068)/150)
[20]:
ir_ilt.invert()
Starting iterations ...
100%|██████████| 100/100 [04:51<00:00, 2.91s/it, Convergence=1.54e-03]
Done.
Plot the results
[21]:
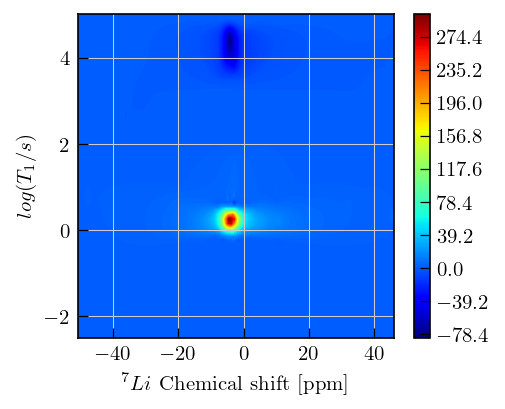
fig,ax = plt.subplots(figsize=(5,4))
x = chemical_shift_ppm[9000:25000:15].flatten() # chemical shift values for the region which was inverted
y = np.log10(ir_ilt.tau[0]).flatten() # ouput sampling vector, T1 values
dist = -ir_ilt.g # relaxation distribution, we plot the negative here since it is an inversion recovery dataset
c = ax.contourf(x,y,dist,cmap='jet',levels=500)
ax.set_xlabel(r'$^{7}Li$'+' Chemical shift [ppm]',fontsize=10)
ax.set_ylabel(r'$log(T_{1}/s)$',fontsize=10)
_=fig.colorbar(c, ax=ax)
[22]:
# we can calculate the T1 value corresponding the the maximum intensity:
dist_max = np.max(dist)
dist_max_index = np.unravel_index(np.argmax(dist),dist.shape)
t1_max = ir_ilt.tau[0][dist_max_index[0]]
print(f"T1 value at max : {t1_max[0]:.2f} s")
T1 value at max : 1.79 s
Note
Inversion recovery curves may exhibit non-zero baselines, which may be interpreted by the algorithm as a slow relaxing component.
This artefact manifests in the obtained relaxation distribution as a negative peak with a long relaxation time constant, as seen above.
You can separate this artifact from the real feature by defining a tau vector which has its maximum farther away from the feature of interest.
This artefact can also be removed using the
sbparameter, as shown in other inversion recovery examples.